How to Use the Search Function
You can easily access the CRISPR-PLANT v2 dataset in three simple steps:
-
Select plant species and gene locus ID of interest:
Check the genome annotation sources used to ensure ID/version compatibility.
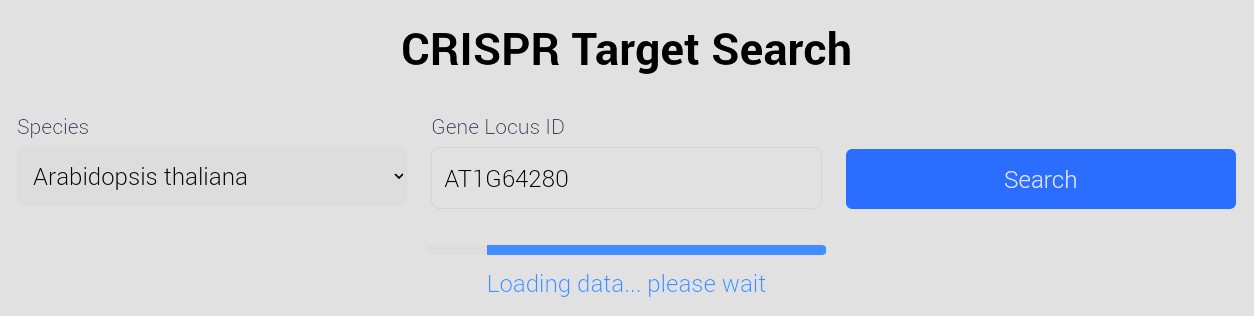
-
View results. Each row includes:
- seqID: Format: Chr:start-end:strand
- class: Specificity group (e.g., A0, B2)
- minMM_GG / minMM_AG: Mismatch count to off-targets
- seq: gRNA spacer sequence (20 nt)
- PAM: Including 7 bp flanking sequence
- strand: Target strand (+ or -)
- location: Genomic feature targeted (exon, intron, etc.)
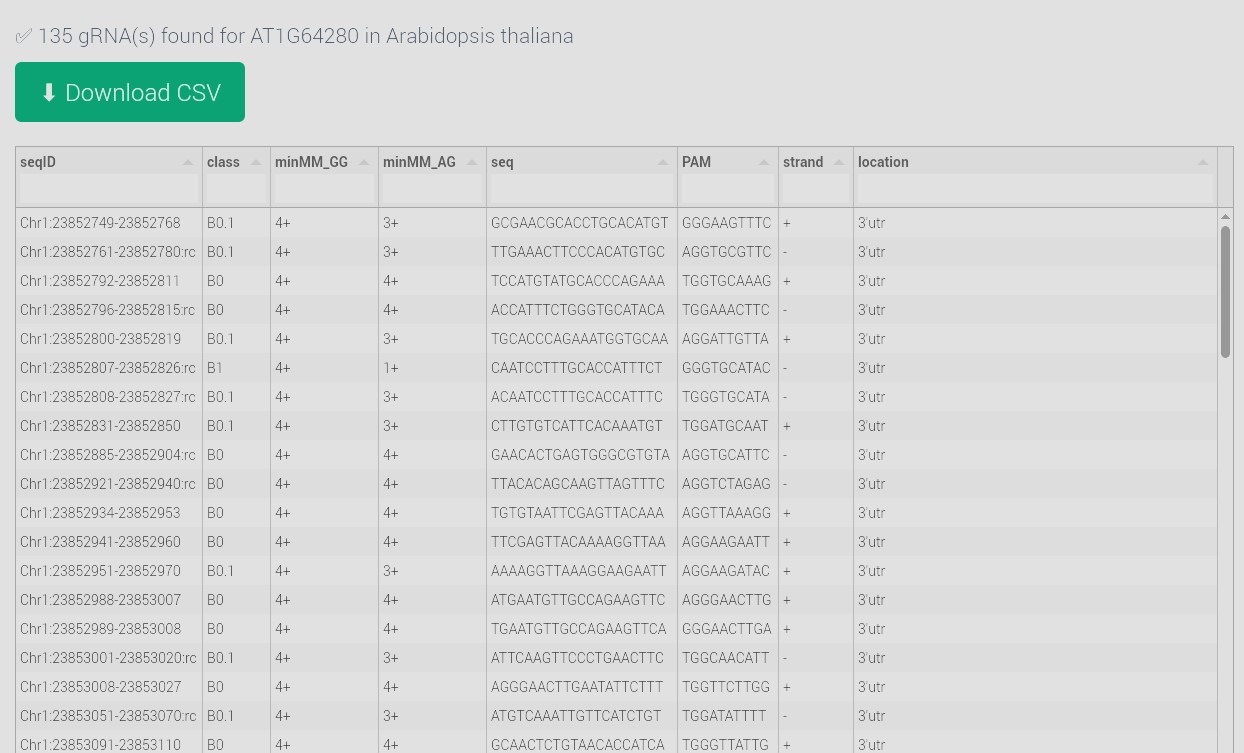
- Press the "Download CSV" button to export your data search.
Spacer Classification
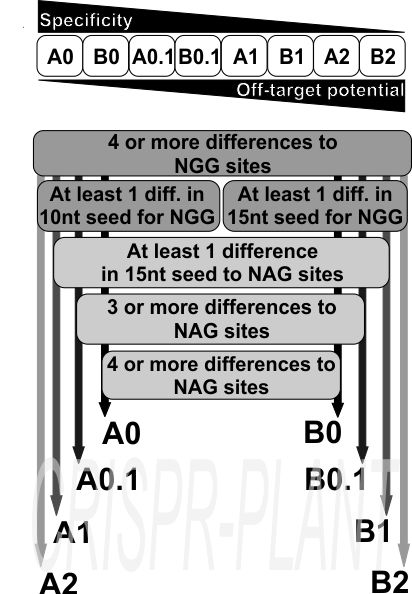
Spacers are classified to reduce off-target risk:
- Class A: 1 mismatch in the 10 bp seed + 3 more elsewhere
- Class B: 1 mismatch in the 15 bp seed + 3 more elsewhere
- Subgroups (0, 0.1, 1, 2) indicate NAG off-target risk
Recommended: A0, A0.1, B0, B0.1 as highly specific spacers.
Learn more in the About Us section.
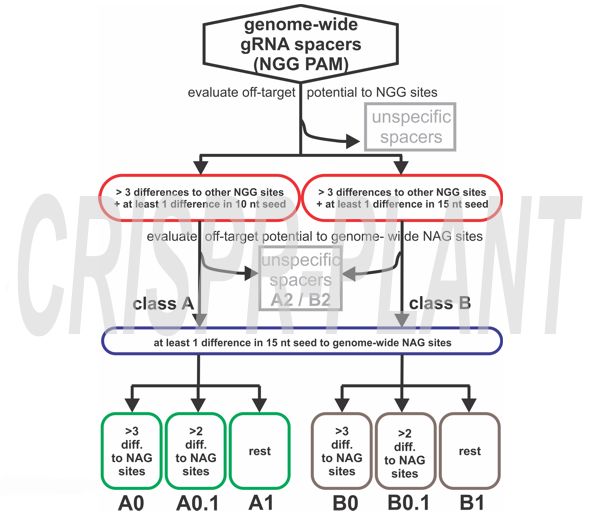
Bioinformatic Pipeline
All NGG and NAG PAM-adjacent spacers are extracted genome-wide and compared. Spacers are grouped by mismatch count and seed region differences. A1–B2 spacers are backups when A0–B0.1 are unavailable.
Genome Annotation Sources
| Species | Group | Genome |
|---|---|---|
| Arabidopsis thaliana | dicot | TAIR10 |
| Medicago truncatula | dicot | Mt4.0v1 |
| Solanum lycopersicum | dicot | ITAG2.3 |
| Glycine max | dicot | Wm82.42.v1 |
| Brachypodium distachyon | monocot | 33v1.0 |
| Oryza sativa | monocot | RGAP Release 7 |
| Sorghum bicolor | monocot | v2 |
Working with a different genome or Cas variant? Clone our pipeline from the GitHub repository and adapt it to your needs.